 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
An 18-month off-road trial will test underground charging systems to juice up on the go and extend driving range
Institute for Highway Safety is known for crash-test safety ratings, but as cars get smarter there's a need to look beyond crashworthiness
It's just a bit shorter than Mt. Everest
Yosemite, Redwood, and other famous parks as they look from outer space
There's a new contender in the century-old quest for perfect, guiltless sweetness: allulose. It's sugar — but in a form that our bodies don't convert into calories. Perfect? Not quite.
Researchers say findings may have important public health implications as vitamin supplements are relatively safe and cost-effective
Free-living songbirds show increased stress hormone levels when nesting under white street lights. But different light spectra may have different physiological effects as this study finds, suggesting that using street lights with specific colour spectra may mitigate effects of light pollution on wildlife
Sarcoptic mange can leave southern hairy-nosed and bare-nosed wombats blind and deaf before eventually killing them
Scientists identify the condition aphantasia, in which people cannot create images in their head
The dust in our homes contains an average of 9,000 different types of fungi and bacteria, a study suggests.
A mosquito can bear up to 23 times its total body weight on each leg, which is crucial for landing on water – the insect's secret is way it stands
Drones are being used to capture video footage that shows construction progress at the Sacramento Kings’ new stadium in California.
8-year-old Harapan joins his brother at an Indonesian breeding sanctuary; fewer than 100 "hairy rhinos" are left in the world
The white Kermode bear of British Columbia is galvanizing First Nations people fighting to protect their homeland
A naturalist cuts through the myths surrounding the invasive plant
Attracting the right species can help get rid of vine-munching insects and allow farmers to cut back on pesticides
It’s becoming increasingly difficult to keep track of online reading material. That’s why physical print-outs sometimes trump a digital copies
Annual vaccinations could be a thing of the past as scientists have successfully tested vaccines on animals infected with different strains of influenza
Websites try to suggest everything from your next best friend to your next best shirt. But are these recommendations a help or a hindrance?
Tropical species with smaller geographical ranges are more likely to die out in a warming climate than those that can adapt by ‘invading’ new regions
Remember winter, when everything was cold and grey? Right now, when all around is lush and green, the contrast couldn’t be greater. But is everything really as it seems? New research shows that we see things differently in winter compared with summer.
Most people think of bacteria as germs, signs of filth, or unwanted bringers of disease. Slowly, that view …
The gloomy octopuses crowded at Jervis Bay, Australia, appear to spit and throw debris such as shell at each other in what could be an intentional use of weapons
Therapies based on hormones that make us more trusting enhance our natural placebo effect – a finding that could alter the way clinical trials are conducted
A new app called Infltr taps into a smartphone's graphics processor to generate filters on the fly, allowing for the perfect shot in one step
Researchers have long struggled to resolve what happens to information when it falls inside a black hole, but the famous physicist says he has a solution
The blind, hairless babies born recently at Washington D.C.'s National Zoo are completely dependent on their mothers—who can sometimes accidentally crush them.
The poop-hoarding insects have an amazingly advanced internal GPS that allows them to navigate by day or night.
Windows 95, the operating system update that changed the way millions of people interacted with their computers, was released 20 years ago today.
Researchers have been using muons to take a peek inside the nuclear reactors in Japan that melted down in 2011. The results could aid the continuing cleanup operations.
. . . More

. . . More
. . . More
Last by Michael Schatz on Feb 26, 2013, 12:13am
The PacBio talk came on the heels of what felt like a warm-up opening act from Jeremy Schmutz of the Hudson Alpha Institute. Schmutz has been working with a start-up that was recently acquired by illumina called . . . More
Yesterday’s first talk was by Russ Altman of Stanford University. Russ has been a leader in the field of pharmacogenomics and he presented his work on developing the Pharmacogenomics Knowledgebase (PharmGKB, pharmGKB.org). He led by saying, “Don’t ever give a talk about a website,” and in his case it was true because WiFi in the conference room was down for the majority of his talk. He urged the crowd to follow along on the website, but only those of us with a cell connection could join him. Russ pointed out the major drawbacks of using GWAS and SNP chips for obtaining information about pharmacoenomic associations and joined pretty much everyone else in saying that the standard t . . . More
Last by Brian Krueger, PhD on Feb 21, 2013, 11:42pm
Despite the tightened biotech purse strings, the event appears from the outset to be extremely well organized. There could be a few more power outlets for those of us carrying a small fortune in lithium powered devices, but I guess I’ll manage. And of course there’s no live streaming or any form of web enabled anything…but I can gripe about that in a long winded and whiny post next week.
The evening opened with a brief introduction with a description of some new meeting changes. This is the biggest AGBT yet with over 800 attendees. A new abstract selection committee was created . . . More
Last by Brian Krueger, PhD on Sep 28, 2012, 5:02pm
However, a quick read of the abstract of the paper yesterday immediately raised a few red flags in my own brain (absolutely full of male cells, by the way). The “discovery” was made using quantitative real-time PCR and only quantitative real-time PCR on a highly repetitive male gene on the Y chromosome. Most people who aren’t scientists don’t . . . More
Last by Brian Krueger, PhD on Sep 08, 2012, 12:05pm

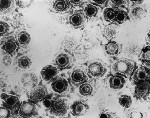
In the lab that I run, we currently work on mutating two different herpesviruses. One of these is Kaposi's Sarcoma Herpesvirus (KSHV) and the other is Murid Herpesvirus 68 (MHV68). Both of these viruses are gammaherpesviruses. In humans, KSHV only really ever becomes a problem in individuals who have a compromised immune system such as those infected with human immunodeficiency virus (HIV). KSHV is an interesting virus because its default program is latency, meaning that once it gets into your cells, it turns itself off and waits for conditions which allow it to grow and take over. This is akin to a bear hibernating in the winter. We do not understand how or w . . . More

One of my test libraries with a perfect library smear. We extract the DNA in between the 200 and 300bp bands for sequencing.
Last by Moderates_Rule on Mar 26, 2013, 11:56am
Last by Brian Krueger, PhD on Mar 17, 2011, 10:17am
. . . More
Last by americanbiotech on Mar 10, 2011, 12:22pm

If money, fame, and science groupies weren't a priority, I'd spend my time and resources trying to find a cure for Cryptocaryon irritans, which is better known in the tropical fish industry as white spot disease, Ich, or Crypto. Crypto is a nasty little bug. It's a protozoan ectoparasite that lodges itself in the epidermis of fish where it grows until it drops off to mature into an Aliens inspired cyst that spits out 300 new little bastard parasites that go on to infect more host fish. In the open ocean, this guy isn't so terrible because its chances of infecting a host are minimal considering the massive amount of water a parasite has to travel through to find a new host. This is a completely different story at an aquaculture facility or in the home aquarium where fish are typically stocked to capacity in relatively small volumes of water. A single cyst can quickly turn into a fish killing epidemic. . . . More
Last by Dr. Girlfriend on Mar 02, 2011, 2:11pm
Anyway, the fall got clogged with trips and holidays. Some of you may remember that back in October I tried to quarantine a yellow longnose butterfly fish and a powder blue tang that both died from a nasty bacterial infection. Given that quarantining fish takes at most 2 months if they get sick, I didn't have enough time to cycle in more fish before Turkey Day! Once Christmas passed, I was working on setting up an office tank for Whitney, so my quarantine system has been occupied with her inhabitants for the past two months... Finally I was able to pick up a new butterfly fish a few weeks ago. . . . More
Last by Thomas Joseph on Jan 04, 2011, 7:47am

. . . More
Human immunodeficiency virus (HIV) was first discovered in the 1980’s when gay men and IV drug users started turning up in hospitals with very odd opportunistic infections like Kaposi’s Sarcoma Herpes virus. These individuals had severely compromised immune systems and the original name given to the condition was gay related immunodeficiency disorder (GRID). The discovery of a viral cause of the disease came in 1983 from the labs of Luc Montagnier (recently won the Nobel Prize for this work) and Robert Gallo (recently didn’t win the Nobel Prize and is kind of pissed about it).
Genetic tests have shown that HIV originated in African monkeys and is related to a similar condition in monkeys called Simian Immunodeficiency Virus (SIV). It is thought that the virus was passed on to humans through the consumption of “bush meat” in sub-saharan . . . More
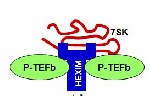
In the fall of 2006, I was finishing up the last few experiments on a project trying to link P53 (a very important protein in cells that helps to prevent cancer) to one of the most important transcriptional activators, P-TEFb. I should probably pause for a second here to give a little bit of background on P-TEFb so that what follows is somewhat understandable. Positive transcription elongation factor b (P-TEFb: Pee Tef bee) is a protein kinase (read as on/off switch) that regulates RNA polymerase II transcription of most genes. RNA polymerase II is the protein responsible for messenger RNA (mRNA) transcription or the process of turning DNA into mRNA. Without mRNA, the transfer form of DNA, proteins cannot be made by the cell, and life does not exist. Essentially, P-TEFb tells RNA polymerase when it’s ok to start making RNA. You can think of P-TEFb as the starter at a track meet, you know, the guy th . . . More
 |
 |





